Problems with BPTT for long sequence classification
Problem Description:
- We have a dataset with long sequences (e.g. 10k time points)
- Each sequence has one label (e.g. ‘Normal Recording ’, ‘Abnormal Recording’)
- We want to train Recurrent Neural Network (RNN) model using Truncated Backpropagation Through Time (Truncated BPTT) to predict the class label.
Design
We create a Toy Dataset with 2 classes. We want to be able to generate long training sequences:
- Sequences filled with 0 for both classes (impossible to overfit)
- Sequences filled with random noise from the same distribution (possible to overfit, impossible to generalize)
- Random sequences repeated twice, once with label 0 and once with label 1 (impossible to overfit)
- Random sequences from different distributions (possible to overfit and possible to generalize)
import numpy as np
class Dataset:
ModeZeros = 'Zeros'
ModeSameRandom = 'SameRandom'
ModeSameDistRandom = 'SameDistRandom'
ModeDiffRandom = 'DiffDistRandom'
def __init__(self, input_size=1, examples_per_class=32, seq_size=10000, mode='Zeros'):
data = []
labels = []
self.seq_size = seq_size
self.index = 0
shape = (examples_per_class, seq_size, input_size)
same = np.random.normal(loc=0.0, scale=1.0, size=shape).astype(np.float32)
for c in range(2):
if mode == self.ModeZeros:
data.append(np.zeros(shape=shape, dtype=np.float32))
elif mode == self.ModeSameDistRandom:
data.append(np.random.normal(loc=0.0, scale=1.0, size=shape).astype(np.float32))
elif mode == self.ModeSameRandom:
data.append(same)
elif mode == self.ModeDiffRandom:
data.append(np.random.normal(loc=0.5 * c, scale=1.0, size=shape).astype(np.float32))
else:
raise RuntimeError('Mode %s not available' % mode)
labels.append(np.ones((examples_per_class, seq_size), dtype=np.int) * c)
self.data = np.concatenate(data)
self.labels = np.concatenate(labels)
def generate_minibatches(self, size):
for i in range(self.seq_size // size):
yield self.data[:, i*size: (i+1)*size, :], self.labels[:, i*size: (i+1)*size]
Model
We create a model and train it using our toy data
- We want to use zeros or random values to create initial state
import torch.nn as nn
import numpy as np
from torch.autograd import Variable
import torch
class RnnBase(nn.Module):
ModeZeros = 'Zeros'
ModeRandom = 'Random'
def __init__(self, input_size, hidden_size, num_layers, output_size):
super().__init__()
self.input_size = input_size
self.hidden_size = hidden_size
self.num_layers = num_layers
self.output_size = output_size
def initial_state(self, mode):
if mode == self.ModeZeros:
return np.zeros((self.num_layers, self.hidden_size), dtype=np.float32)
elif mode == self.ModeRandom:
random_state = np.random.normal(0, 1.0, (self.num_layers, self.hidden_size))
return np.clip(random_state, -1, 1).astype(dtype=np.float32)
else:
raise RuntimeError('No mode %s' % mode)
def initial_states(self, mode, samples=64):
states = [self.initial_state(mode) for _ in range(samples)]
states = np.stack(states)
states = np.swapaxes(states, 1, 0)
states = Variable(torch.from_numpy(states), requires_grad=False)
return states
class SimpleRNN(RnnBase):
def __init__(self, **kwargs):
super().__init__(**kwargs)
self.rnn = nn.GRU(input_size=self.input_size, hidden_size=self.hidden_size, num_layers=self.num_layers,
batch_first=True)
self.fc = nn.Linear(in_features=self.hidden_size, out_features=self.output_size, bias=True)
def forward(self, x, hidden):
lstm_out, hidden = self.rnn(x, hidden)
lstm_out = lstm_out.contiguous()
fc_out = self.fc(lstm_out.view(lstm_out.size(0) * lstm_out.size(1), lstm_out.size(2)))
fc_out = fc_out.view(lstm_out.size(0), lstm_out.size(1), fc_out.size(1))
return fc_out, hidden
Training
And we train the model:
In each iteration we pass chunks of length ‘bptt_size’ from each training sequence. If our sequences are 10k steps long and ‘bptt_size’ is 100 then one epoch corresponds to 100 updates.
First we pass the trainig data applying gradient update in each iteration and we record the training loss. Then, we pass the same trainign data but now without any parameter updates.
def main(cuda, learning_rate, weight_decay, input_size, examples_per_class, epochs, bptt_size,
hidden_size, num_layers, initial_state_type, data_type):
data = Dataset(input_size=input_size, examples_per_class=examples_per_class, seq_size=10000,
mode=data_type)
model = SimpleRNN(input_size=input_size, hidden_size=hidden_size, num_layers=num_layers,
output_size=2)
plotter = Plotter()
plotter.plot_sequences(data.data, 'input.png')
criterion = nn.CrossEntropyLoss()
if cuda:
model = model.cuda()
criterion = criterion.cuda()
optimizer = torch.optim.Adam(model.parameters(), lr=learning_rate, weight_decay=weight_decay)
losses_per_epoch = {True: [], False: []}
losses_last_epoch = {True: [], False: []}
for epoch in range(epochs):
for update in [True, False]:
hidden = model.initial_states(mode=initial_state_type, samples=examples_per_class*2)
if cuda:
try:
hidden = hidden.cuda()
except AttributeError:
hidden = hidden[0].cuda(), hidden[1].cuda()
losses = []
for batch, labels in data.generate_minibatches(size=bptt_size):
batch = Variable(torch.from_numpy(batch))
labels = Variable(torch.from_numpy(labels))
if cuda:
batch = batch.cuda()
labels = labels.cuda()
hidden = repackage_hidden(hidden)
outputs, hidden = model(batch, hidden)
last = True
if last:
training_outputs = outputs[:, -1, :]
training_labels = labels[:, -1]
else:
outputs_num = outputs.size()[-1]
training_outputs = outputs.view(-1, outputs_num)
training_labels = labels.view(-1)
loss = criterion(training_outputs, training_labels)
if update:
# Backward pass
optimizer.zero_grad()
loss.backward()
torch.nn.utils.clip_grad_norm(model.parameters(), 1)
optimizer.step()
l = loss.cpu().data.numpy()[0]
losses.append(l)
if epoch == epochs - 1:
losses_last_epoch[update].append(l)
l = sum(losses)/len(losses)
print('Epoch %d, Update: %s, Loss: %g' % (epoch, update, l))
losses_per_epoch[update].append(l)
if epoch > 0:
plotter.plot_losses(losses_per_epoch, 'losses.png')
plotter.plot_losses(losses_last_epoch, 'losses_last_epoch.png')
Experiment
- We train a simple RNN model with one layer (512 neurons) using random input data.
- We use the setting where the same sequence appears twice in the dataset, once with label “0” and once with label “1”. There should be no way for the model to differentiate between both.
- In the first iteration we feed different random states for each example.
- In subsequent iterations we simply forward hidden states from the previous iteration.
python main.py --hidden_size 512 --epochs 25 --bptt_size 10 --num_layers 1 --initial_state_type Random --learning_rate 0.002 --data_type SameRandom --examples_per_class 32
Results might come surprising. They were for me at the beginning when I used real world dataset with similar properties to the toy dataset (long sequences with just one label for the whole sequence).
Loss per epoch:
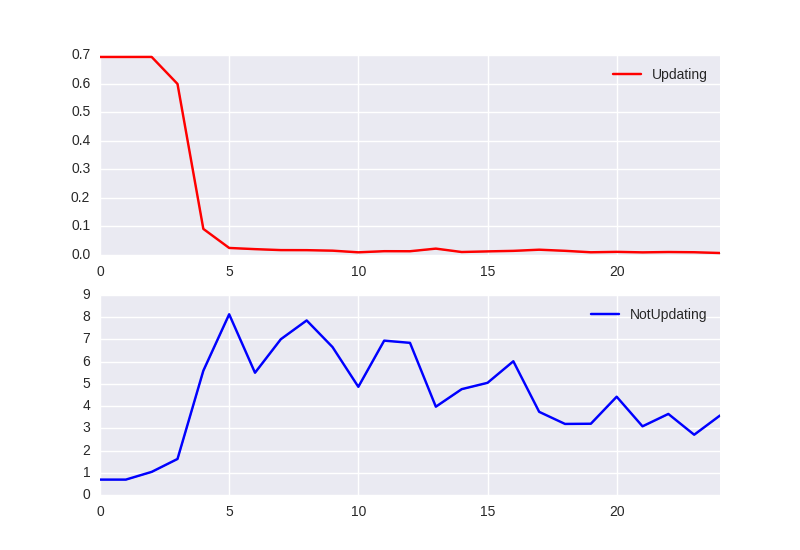
When we pass training data and apply updates to the weights, network is able to correctly classify sequences. However, when we use this network and pass the same data again but now without updates it gives random predictions.
What happens:
Instead of looking at the inputs, network learns to classify random hidden state which is used as an initial state. Important thing to notice is that in each epoch network needs to learn it again as different random states are used.
On the next plot we show how the loss changes during the last epoch:
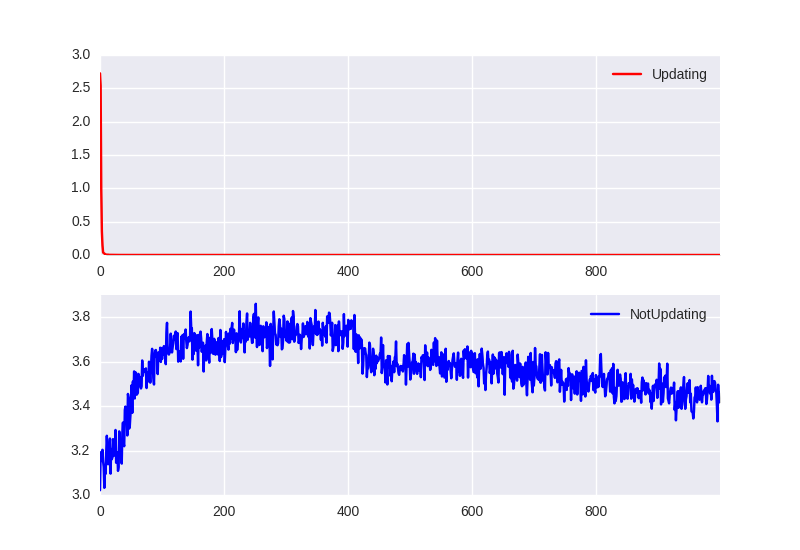
We see that network needs just a cuple of training iterations to properly classify the new random state (Fig. 2). Network develops an ability to quickly learn to classify new hidden state. Interestingly it needs couple of epochs at the beginning to achieve this property (Fig. 1).
We used a simple setting where ‘bptt_size’ is 10 but the same behavior was observed on real word data with thousands of training examples where ‘bptt_size’ was 1000 and training sequences were longer than 100k.
Take out message
Be careful when you train using Truncated BPTT on long sequences where each subsequence has the same label. What might happen is that network will develop the ability to quickly learn to classify new random hidden states in each epoch. Even if you fill initial states with 0, after first iteration they will change based on the input data, so the problem still preserves (you can run experiments with –data_type SameDistRandom and –initial_state_type Zeros to see this behavior).